|
0 Comments
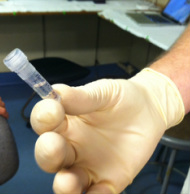 Gold! Gold! Hard to believe but this tiny bottle contains genetic information that will allow us to characterize the diversity of the microbial community at Station ALOHA. This was the final product from 5 days of sampling and many hours filtering, extracting, priming, amplifying and pipetting! We took water samples from the surface of the water all the way through the water column. We are interested in determining the diversity of microbial organisms at Station Aloha. We used PCR (polymerase chain reaction) to amplify pieces of DNA across several orders of magnitude. Each sample received a specific primer which serves as "tag" so we can link the microbial community to a depth and time of day they were collected. Primers are short DNA fragments that contain sequences complementary to the target region along the DNA polymerase. We checked to see whether the PCR generated the anticipated DNA fragments (sometimes called amplicons) using a process called agarose gel electrophoresis, which separates the PCR products based on size. A DNA ladder containing DNA fragments of known sizes is run alongside the PCR products. Once we determined that the PCR worked for each sample we then conducted a procedure that pooled all of the samples together and we ended up with one small bottle. It is truly mind blowing to consider how much data and information is contained in such a small volume! 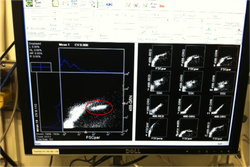 Prochlorococcus community circled in red. Prochlorococcus community circled in red. Flow cytometry is a laser based biophysical technology used for cell counting and sorting. It is used widely in the medical field to analyze blood samples but over the past few decades it has become an important instrument used in marine biology to determine microbial community structure and abundances. Flow cytometry suspends cells in a stream of fluid that then passes by an electronic detector. The analysis can characterize the physical and chemical attributes of up to thousands of particles per second. At Station Aloha and other low nutrient regions of the ocean, a genus of photosynthetic microorganisms called prochlorococcus are abundant and can be identified and quantified using flow cytometry (see picture). Prochlorococcus are incredibly small (0.6um) cyanobacteria that are the most abundant photosynthetic organisms on earth. One milliliter of seawater may contain up to 100,000 cells of these organisms. Due to their small size Prochlorococcus have a high surface to volume ratio which gives them a competitive advantage in low nutrient waters. They can also grow at very low light conditions. Despite their ecological importance they were not discovered until 1986 by Sallie W. (Penny) Chisholm (Massachusetts Institute of Technology) and other collaborators. The discovery was made using flow cytometry methods in the Sargasso Sea.  SQF getting ready to recover the array. SQF getting ready to recover the array. Thursday was an awesome day! I love working on the deck. For the whole day our group was responsible for the deployment and recovery of everything put into the water. This included all of the CTD/niskin rosette casts, the productivity array, and optical instrument casts. The day started at 11:30pm (the night before)! Our first cast was at midnight and it included 3 different optical instruments plus a small CTD that were attached to the same metal cage. The optical instruments included a LISST (Laser In Situ Scattering Transmissometry), AC9, and ACplus. These are active optical instruments which means that they analyze the optical properties of the water using laser and light rays opposed to using light from the sun. This is why we can do the cast in the middle of the night. We also deployed a productivity array which contained bottles of sea water spiked with the radio isotope 14C and other bottles spiked with tritiated leucine (an amino acid with the radio isotope 3H). These bottles were placed on a line at different depths and then put into the water at sunrise and then floated around attached to buoys and an Argos transmitter. At sunset we tracked down the array and hauled it back on deck. The samples were then analyzed to determine the rate of CO2 uptake (primary productivity) using the 14C label and also the rate of protein synthesis /bacterial productivity using the tritiated leucine label.  CTD and niskin rosette. CTD and niskin rosette. Aloha! Today is Saturday 6.15.2013, day 5 of our research cruise. It has been an exciting week of science! Due to slow/inconsistent internet it has been difficult to update this blog and we have done a lot so I have tons to write about! On Wednesday 6.12.2013 my student group ("The Kraken", self-named:) worked on analyzing water samples collected at multiple depths between the surface of the water down to 1000 meters using the niskin sampling tubes on the CTD rosette (see picture). We took samples to measure important basic water quality parameters such as dissolved inorganic nutrients, dissolved oxygen, particulate carbon, nitrogen and phosphorous, and ATP. The ATP analysis was particularly interesting. ATP (Adenosine-5' triphosphate), is a coenzyme that transports energy within cells for metabolism. It can be thought of as the "molecular unit of currency". Since ATP is found in and around all living organisms, measuring the total concentration (amount per volume) of ATP in our water samples provides a measure of total microbial biomass. The analysis for quantifying ATP is super cool...After filtering our water sample onto a small filter, we place it into a tube filled with a boiling base solution. This step breaks up the cells and extracts the ATP. After the extraction we add "firefly dust" which contains an enzyme (luciferase) and subtrate (luciferin) extracted from fireflies! The reaction produces light and the amount of light produced is directly proportional to the amount of ATP and therefore the amount of living organisms in the sample! The reaction is catalyzed by the enzyme (luciferase) simplified to this: luciferin + ATP --> oxyluciferin + AMP + light  SQF leaving the port in Honolulu. SQF leaving the port in Honolulu. First day of our research cruise to Station Aloha. We got underway at 9am this morning and we have been making our way up the western coast of Oahu. We just past the most western point of the island and are heading north leaving the land behind us. What a beautiful day! I wish I could take a picture that captured how blue the water is! Our day started with safety training and drills with the captain of the R/V Kilo Moana. Later in the morning we did a test CTD deployment and cast. The CTD is a submersible instrument that measures Conductivity (salinity), Temperature, and Depth as it is lowered through the water column. Attached to the CTD are other sensors that measure light, water clarity, dissolved oxygen, nitrate, and fluorescence (measure of chlorophyll a which is a proxy for phytoplankton biomass). After the cast, my group ("The Kraken") prepped the sediment trap tubes (see the second pic). These will deployed tonight at midnight and they will be collected in 5 days. They will capture the "stuff" (sometimes referred to as marine snow) that falls through the water column. We can measure the carbon and nutrient content of this material and it will give us a sense of what type of material is being transported from the upper part of the water column to the lower depths. This is important because the marine snow is a food source for many sea creatures. A very small portion of these particles may make it to the bottom of the sea where it may be consumed or buried. The movement of carbon (found in marine snow) from the surface of the ocean to the interior of the sea is called the biological pump. This pump plays an important role in global carbon cycle. Today we spent moving supplies to the R/V Kilo Moana and setting up the labs. Our new home for the next 10 days! The research vessel (R/V) Kilo Moana is a 186 foot, twin hull vessel. Tomorrow morning we will steam out to Station Aloha which is about 115 kilometers (~71.5 miles) off the North Shore coast of Oahu. We will leave from the Univ. of Hawaii Marine Center in Honolulu and steam around Oahu and up to Station Aloha. It should take us about 10 hours to get to station. Once we are on station we will begin our measurements and rotations. All of the C-MORE students are broken up into 5 groups of 3 students and we will rotate between 5 different types of measurements; Biomass, Metabolism, Diversity and Enumeration of the Microbial Community, Flow Cytometry, Optics and deck work. I will describe these in more detail once I do the rotations. Now it is time for me to get some sleep before the departure tomorrow morning at 7am!
Aloha from the middle of the Pacific Ocean! This week concluded Part I of the summer Microbial Ecology course I am taking at the world renowned Center for Microbial Ecology Research and Education (C-MORE) at the University of Hawaii at Manoa. It is hard to believe that I have been in Hawaii for 2 weeks! The course is taught by a group of local and visiting faculty directed by Dave Karl.
The first week was broadly focused on "The Ocean as a Habitat". The visiting faculty included Ginger Armbrust (Univ. of Washington), John Cullen (Dalhousie Univ.), Ken Johnson (MBARI) and Dan Repeta (WHOI). We had 3 to 4 lectures/discussions per day with topics including a general introduction and history of microbial ecology; nutrients, light and primary productivity; nutrient cycling; observations of biogeochemistry with robotics; modeling; functional and taxonomic diversity of phytoplankton; and carbon cycling. It was an incredibly dynamic and interesting week. Check out this review on signaling interactions between diatoms and bacteria: http://mmbr.asm.org/content/76/3/667.short And also check out the awesome MBARI database of data from deployed floats across the ocean: http://www.mbari.org/chemsensor/floatviz.htm The second week of the class was titled "From Genomes to Biomes" and was led by Ed DeLong (MIT). The lectures covered general marine microbial 'omics and the cutting edge methods and technology being used. The visiting faculty included Eric Allen (Scripps Institution, UCSD), Elizabeth Costello (Stanford Univ.), Scott Gifford (MIT) and Nikos Krypides (JGI, DOE). We also had lectures from local C-MORE faculty Mike Rappe, Grieg Stewart and Benedetto Barone. We were introduced to genomic software tools including IMG/ER, CAMERA, and Qiime. What a full and exciting couple of weeks! Now we will temporarily leave C-MORE Hale (LEED Platinum certified) and head to the big blue! In Part II of the course we will conduct research on a research cruise to Station ALOHA (A Long-Term Oligotrophic Habitat Assessment, 22deg 45'N, 158deg 00'W). We will be at sea for a total of 10 days. This will be my first open ocean cruise... I can't wait. |
Sarah Q. FosterSarah is a 2nd year Ph.D. student in the Fulweiler Lab. This blog documents her experience taking a summer course "Microbial Oceanography: From genome to biome" at C-MORE at the University of Hawai'i at Manoa. ArchivesCategories |
















 RSS Feed
RSS Feed